-
Notifications
You must be signed in to change notification settings - Fork 6
Decision Tree in Python : Metaprotein
We have implemented the following criteria to split nodes in our Decision Trees for comparison of accuracy
- Gini Index
- Information Gain (Entropy)
Metaproteins as rows and, patient data of three types with samples from each being tested for the presence of metaproteins in columns (along with metaprotein demographics)
To suit our decision tree model, we removed the demographic columns from the dataset and have transposed the data frame to turn metaproteins into columns/variables & patients as rows.
We created a class label "Patient type" which has 3 factors - C, UC & CD
We have taken half (1/2) of our Metaprotein Dataset to be used as Training Dataset & (1/2) to be used as Testing Dataset
- depth = 2
- leaf nodes = 4
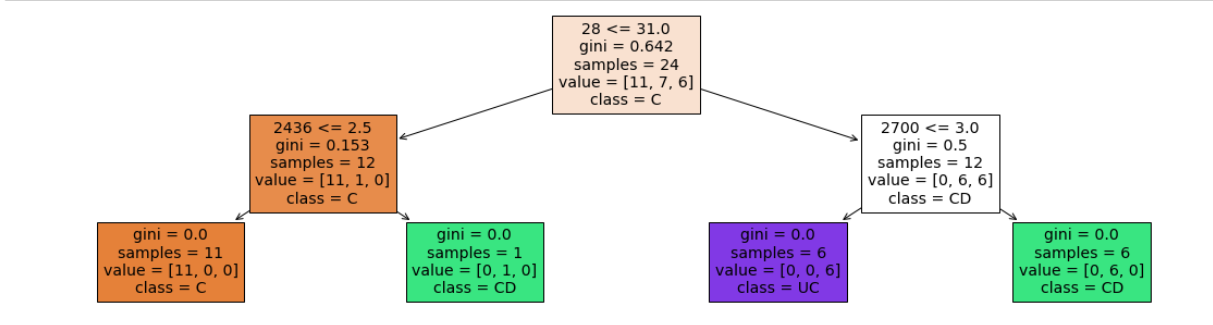
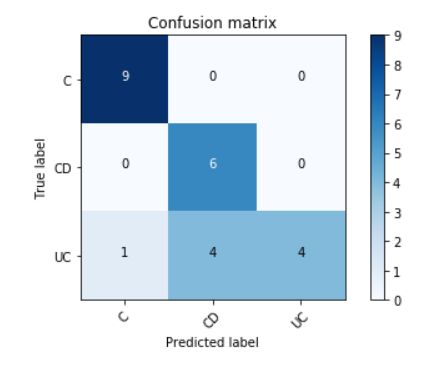
Accuracy = 19/24 = 79.16 %
- depth = 2
- leaf nodes = 4
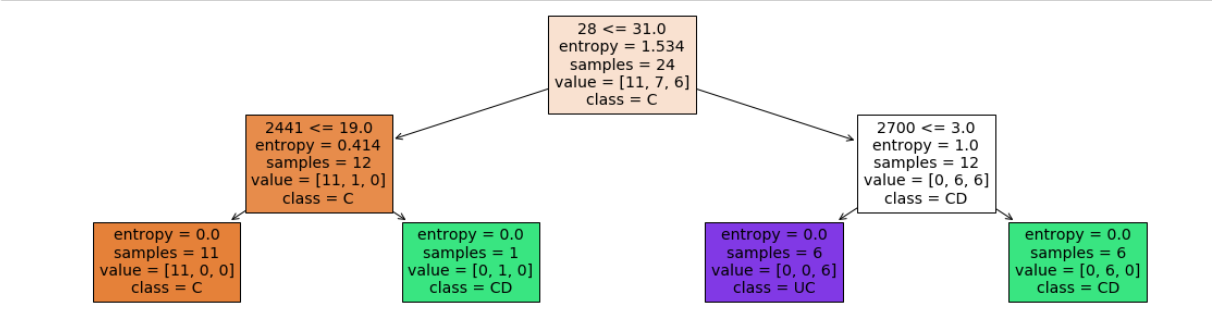
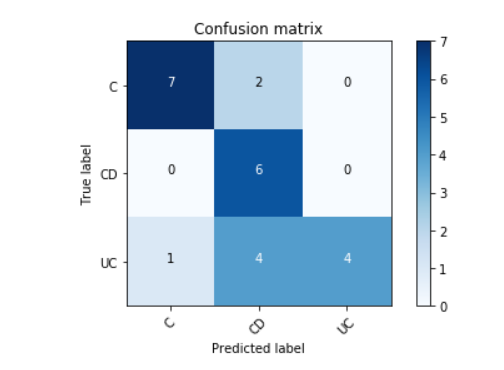
Accuracy = 17/24 = 70.83 %
A tree that is too large risks overfitting the training data and poorly generalizing to new samples. We realized the necessity of pruning the decision trees by tuning parameters namely, "max_depth" and "max_leaf_nodes".
- depth = 1
- leaf nodes = 2
Accuracy = 54.1 %
- depth = 2
- leaf nodes = 3
Accuracy = 70.8 %