-
Notifications
You must be signed in to change notification settings - Fork 6
Decision Tree in Python : Metaprotein
We have implemented the following criteria to split nodes in our Decision Trees for comparison of accuracy
- Gini Index
- Information Gain (Entropy)
Metaproteins as rows and, patient data of three types with samples from each being tested for the presence of metaproteins in columns (along with metaprotein demographics)
To suit our decision tree model, we removed the demographic columns from the dataset and have transposed the data frame to turn metaproteins into columns/variables & patients as rows.
We created a class label "Patient type" which has 3 factors - C, UC & CD
The metaprotein dataset has 2970 variables and 48 instances after the removal of demographic information and the addition of the class label. A vast majority of these variables are completely filled with 0 and they have an adverse effect on the accuracy of decision tree algorithms. We have chosen the 50 most abundant variables in the dataset and created our algorithms with them.
We have taken half (1/2) of our Metaprotein Dataset to be used as Training Dataset & (1/2) to be used as Testing Dataset
- depth = 4
- leaf nodes = 5
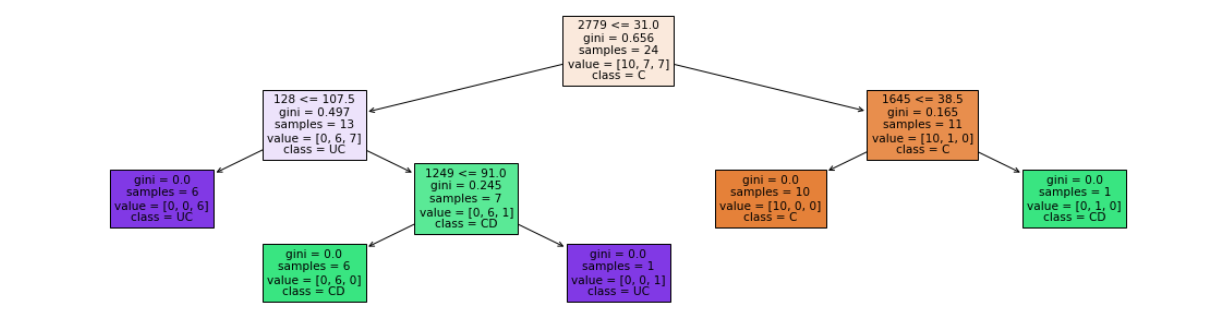
Accuracy = 75 %
- depth = 4
- leaf nodes = 5
Accuracy = 66.66666667 %