|
| 1 | +# cellpose-plgs |
| 2 | + |
| 3 | +**Path:** https://github.com/Image-Py/cellpose-plgs |
| 4 | + |
| 5 | +**Version:** 0.1 |
| 6 | + |
| 7 | +**Author:** YXDragon, Carsen-Stringer |
| 8 | + |
| 9 | +**Email:** yxdragon@imagepy.org |
| 10 | + |
| 11 | +**Keyword:** cellpose, segment, unet |
| 12 | + |
| 13 | +**Description:** cellpose is a generalist algorithm for cell and nucleus segmentation, this is a cellpose plugin for imagepy. |
| 14 | + |
| 15 | + |
| 16 | + |
| 17 | +## Document |
| 18 | +### Install |
| 19 | + |
| 20 | +there are two method to install cellpose-plgs, Install Plugins or use Plugins Manager |
| 21 | + |
| 22 | +1. **Menus: Plugins > Install > Install Plugins** then input https://github.com/Image-Py/cellpose-plgs |
| 23 | + |
| 24 | + 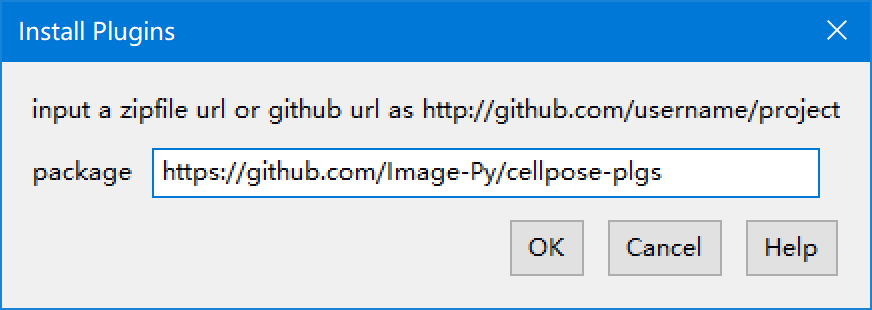 |
| 25 | + |
| 26 | + |
| 27 | + |
| 28 | +2. **Menus: Plugins > Manager > Plugins Manager**, befor you open the manager, you should update the software, (to getting the newest plugins catlog). for git version, please pull, for release version, please use *Menus: Plugins > Updata Software*. |
| 29 | + |
| 30 | + 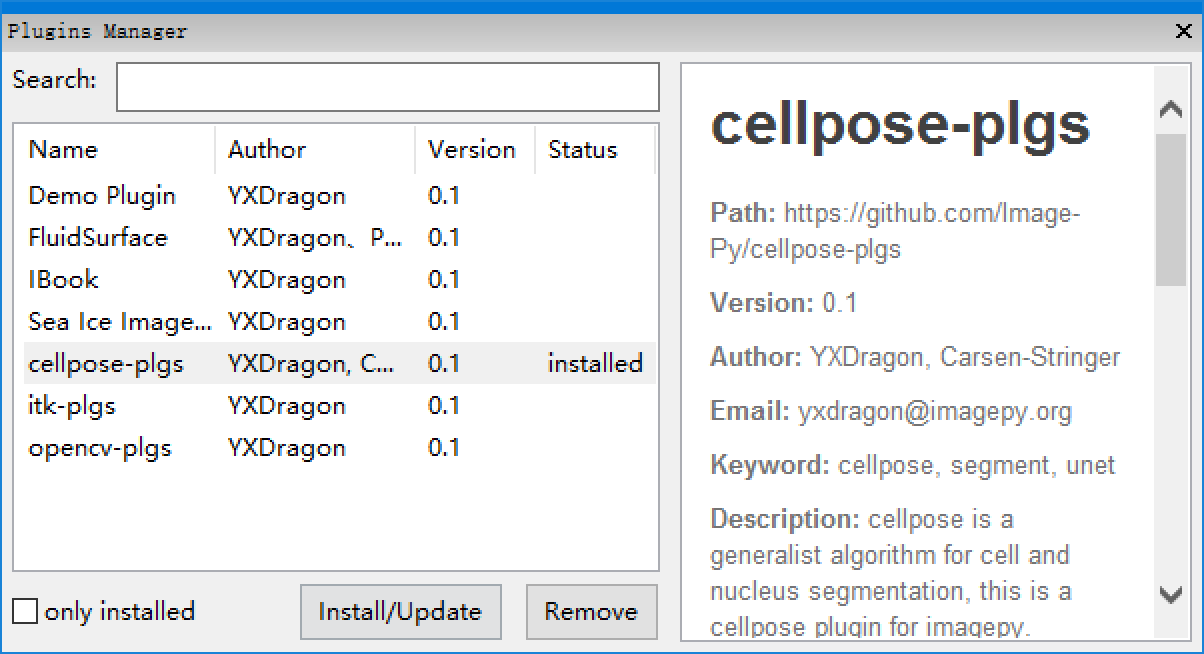 |
| 31 | + |
| 32 | +when the cellpose-plgs is installed seccessfully, The cellpose would appears below the **Plugins** menus. |
| 33 | + |
| 34 | + |
| 35 | + |
| 36 | +### Usage |
| 37 | + |
| 38 | +**Menus: File > Open** to Open a Image, you can also use **Menus: File > Import > Import Sequence** to open a sequence. Then **Menus: Plugins > Cell Pose > Cell Pose Eval**, you would got a parameter dialog, Setting the parameter, then OK. |
| 39 | + |
| 40 | + |
| 41 | + |
| 42 | +**Parameters:** |
| 43 | + |
| 44 | +**model:** select model |
| 45 | + |
| 46 | +**cytoplasm:** select the cytomplasm channel. 0: gray, 1:red, 2:green, 3:blue |
| 47 | + |
| 48 | +**nucleus:** select the nucleus channel, if there is no, select 0. |
| 49 | + |
| 50 | +**show color flow:** the color flow result would be shown when checked. |
| 51 | + |
| 52 | +**show diams tabel:** the diams result would be shown when checked. |
| 53 | + |
| 54 | +**slice:** process the current image or all images when it is a sequence. |
| 55 | + |
| 56 | +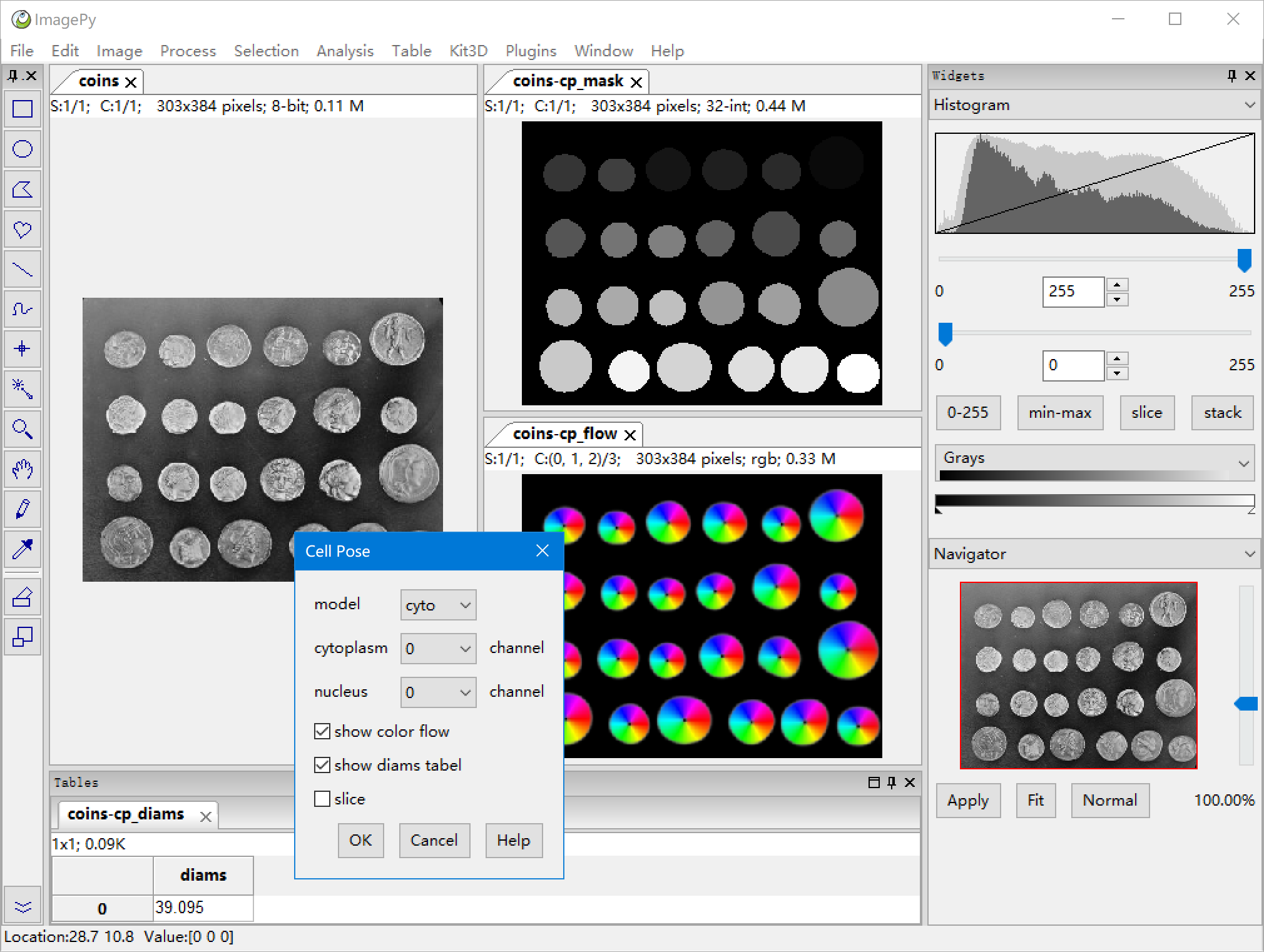 |
| 57 | + |
| 58 | + |
| 59 | + |
| 60 | +### Analysis |
| 61 | + |
| 62 | +**Menus: Analysis > Region Analysis > Geometry Analysis** do a region analysis. And we can do many other things in ImagePy, such as set a colormap, or overlay the mask on the original image. |
| 63 | + |
| 64 | +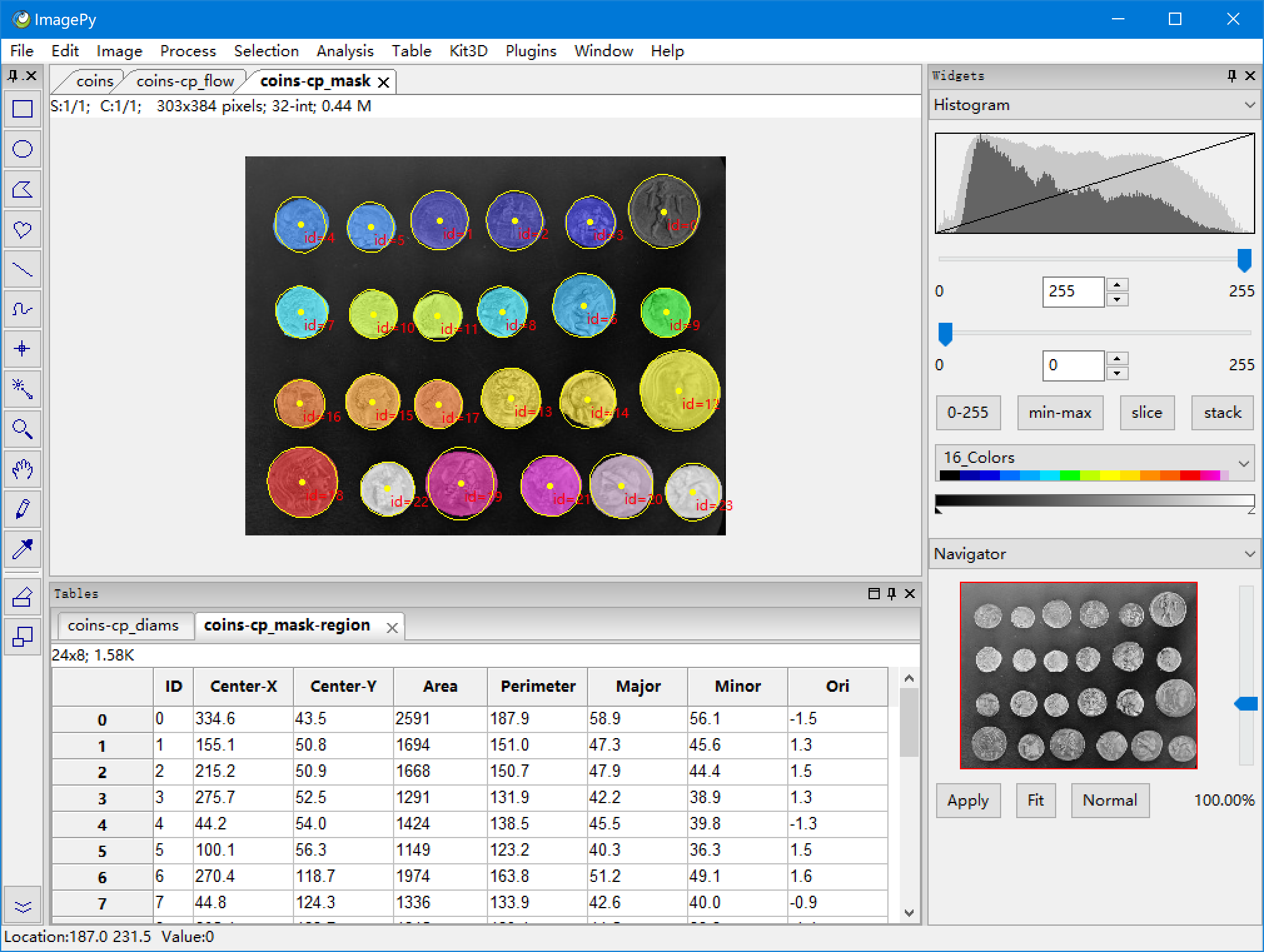 |
| 65 | + |
| 66 | + |
| 67 | + |
| 68 | +### Reference |
| 69 | + |
| 70 | +Thanks for CellPose and the supporting from Carsen Stringer and Marius Pachitariu. More detail about CellPose please read the [paper](https://t.co/4HFsxDezAP?amp=1) or watch the [talk](https://t.co/JChCsTD0SK?amp=1). |
0 commit comments