|
| 1 | +## Automatical parameters optimization |
| 2 | +As we have developed five different metrics, the reconstruction result could be evaluated quantitatively. So we write a script to search for the best parameters automatically. |
| 3 | +Simulated Annealing is used to find the global optimal result, and the loss value in each iteration is calculated by averaging the precision and recall of ssd metric. |
| 4 | + |
| 5 | +### 1. Locate the optimization script, data and neutu |
| 6 | +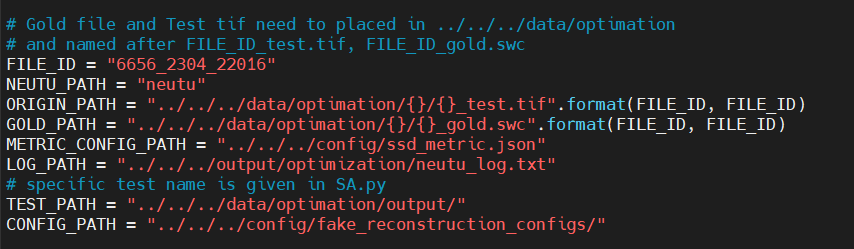 |
| 7 | +#### 1.1 Tools and configs of tools: |
| 8 | +##### NEUTU_PATH: |
| 9 | +The reconstruction process relies on neutu(https://github.com/janelia-flyem/NeuTu), the path executable neutu program need to be list. |
| 10 | +In the example, neutu has been added to enviroment variable so only a "neutu" is enough. |
| 11 | +##### METRIC_CONFIG_PATH: |
| 12 | +Metric methods such as SSD requires configs in json format too. In most case default configs are enough. |
| 13 | +##### LOG_PATH |
| 14 | +The process of tracing in Neutu produce quite a lot of log data prevent us from getting the log about optimization. So we redirect these log to files in this path. |
| 15 | +#### 1.2 Input |
| 16 | +The input of contains three parts known as: |
| 17 | +##### ORIGIN_PATH: |
| 18 | +origin neuron image in TIFF format |
| 19 | +##### GOLD_PATH: |
| 20 | +gold standard reconstruction result in SWC format |
| 21 | +##### CONFIG_PATH: |
| 22 | +path of initial parameters of reconstruction |
| 23 | + |
| 24 | +You can create a folder named by FILE_ID in ($Pyneval)/data/optimization/ and name the ORIGIN tiff with FILE_ID_test.tif, name the GOLD swc with FILE_ID_gold.swc, |
| 25 | +and leave initial parameters of reconstruction with default value. Of course you can ignore FILE_ID and change these PATH as you like. Only make sure you can understand the meaning of these path. |
| 26 | + |
| 27 | +#### 1.3 OUTPUT |
| 28 | +This script provides two output: |
| 29 | +##### optimal parameters |
| 30 | +the optimal parameters among all the iterations are save in the following path |
| 31 | +in ($PyNeval)/config/fake_reconstruction_configs/best_x_{FILE_ID}.json |
| 32 | + |
| 33 | +##### optimal reconstruction result |
| 34 | +The optimal reconstruction will be saved in: |
| 35 | +TEST_PATH/FILE_ID.swc |
| 36 | + |
| 37 | +### 2. run the script |
| 38 | +Once you have confirmed all the path above, run: |
| 39 | +```python optimize.py``` |
| 40 | +in ($Pyneval)/pyneval/tools/optimize/ and waiting for results |
| 41 | + |
| 42 | + |
| 43 | +### 3. Adjusting parameters of SA |
| 44 | +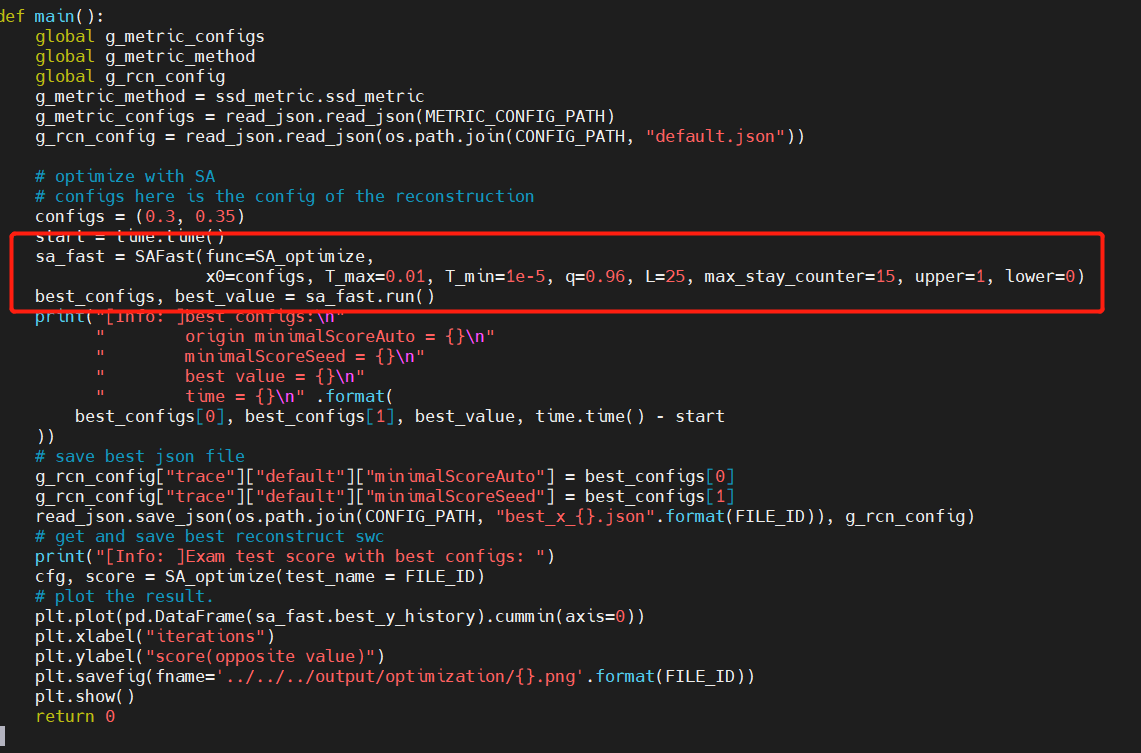 |
| 45 | +All the configs in SA can be adjust in the main function |
| 46 | +##### T_max: initial temperature |
| 47 | +##### T_min: minimum temperature, script stops if temperature is lower then T_min |
| 48 | +##### q:descending rate of T |
| 49 | +##### L: number of random loops under each iteration |
| 50 | +##### max_stay_counter: script stop if no new optimal value is found after max_stay_counter times of temperature descension. |
| 51 | +##### upper, lower: resize the step of random movement in each iteration |
0 commit comments